The ordertrans() function in the
hyper2 package
Robin K. S. Hankin
Source:vignettes/ordertrans.Rmd
ordertrans.Rmd
ordertrans## function (x, players)
## {
## if (missing(players)) {
## return(x[order(names(x))])
## }
## else {
## if (is.hyper2(players) || is.hyper3(players)) {
## players <- pnames(players)
## }
## }
## stopifnot(length(x) == length(players))
## stopifnot(all(sort(names(x)) == sort(players)))
## stopifnot(all(table(names(x)) == 1))
## x[apply(outer(players, names(x), `==`), 1, which)]
## }To cite the hyper2 package in publications, please use
Hankin (2017). The
ordertrans() function can be difficult to understand and
this short document provides some sensible use-cases. The manpage
provides a very simple example, but here we are going to use an even
simpler example:
x <- c(d=2,a=3,b=1,c=4)
x## d a b c
## 2 3 1 4In the above, object x is a named vector with elements
seq_along(x) in some order. It means that competitor
d came second, competitor a came third,
b came first and c came fourth. Technically
x is an order vector because it answers the question “where
did a particular competitor come?” However, it might equally be a rank
vector because it answers the question “who came first? who came
second?” (see also the discussion at rrank.Rd).
But note that x is not helpfully structured to answer
either of these questions: you have to go searching through the names or
the ranks respectively—both of which may appear in any order—to find the
competitor or rank you are interested in. To find the rank vector (that
is, who came first, second etc), one would use sort():
sort(x)## b d a c
## 1 2 3 4But this is suboptimal to find the order vector [“where did a
particular competitor come?”]. For the order vector, we want to
rearrange the elements of x so that the names are in
alphabetical order. That way we can see straightaway where competitor
a placed (in this case, third). This nontrivial task is
accomplished by function ordertrans():
o <- ordertrans(x) # by default, sorts names() into alphabetical order
o## a b c d
## 3 1 4 2Observe that objects x and o are equal in
the sense that they are a rearrangement of one another:
## [1] TRUE## [1] TRUEOne consequence of this is that the resulting Plackett-Luce support functions will be the same:
(Sx <- ordervec2supp(x))## log( a * (a + b + c + d)^-1 * (a + c)^-1 * (a + c + d)^-1 * b * d)
(So <- ordervec2supp(o))## log( a * (a + b + c + d)^-1 * (a + c)^-1 * (a + c + d)^-1 * b * d)Note carefully that the two support functions above can be
mathematically identical but not formally identical()
because neither the order of the brackets, nor the order of terms within
a bracket, is defined. They look the same on my system but YMMV. It is
possible that the two support functions might appear to be different
even though they are mathematically the same. We can verify that the two
are mathematically identical using package idiom ==:
Sx==So## [1] TRUEAnother use-case
How about this:
(x <- c(d=2, a=3, c=4, b=3, e=6))## d a c b e
## 2 3 4 3 6
(y <- c(e=3, c=2, a=4, b=5, d=1))## e c a b d
## 3 2 4 5 1
x+y## d a c b e
## 5 5 8 8 7Above, observe that R is behaving as intended
but is potentially confusing. Take the first element. This has value
2+5=7 and name d from the name of element 1 of
x. But it would be reasonable to ask for the first element
to be the sum of element d of x and element
d of y, which would be 2+1=3.
This can be done with ordertrans():
ordertrans(x) + ordertrans(y)## a b c d e
## 7 8 6 3 9See how ordertrans() has rearranged both x
and y so that the names are in alphabetical order.
But this is not perfect. Consider:
(z <- c(f=3, g=2, h=4, a=5, b=1)) # names NOT letters[1:5]## f g h a b
## 3 2 4 5 1
ordertrans(x) + ordertrans(z) # arguably not well-defined## a b c d e
## 8 4 7 4 10In this case the result is arguably incorrect.
Skating
Let us consider the skating dataset and use ordertrans()
to study it (note that the skating dataset is analysed in more depth in
skating.Rmd).
skating_table## An ordertable:
## J1 J2 J3 J4 J5 J6 J7 J8 J9
## hughes 1 4 3 4 1 2 1 1 1
## slutskaya 3 1 1 1 4 1 2 3 2
## kwan 2 3 2 2 2 3 3 2 3
## cohen 5 2 4 3 3 4 4 4 4
## suguri 4 8 5 5 5 7 5 5 5
## butyrskaya 6 5 8 7 12 5 8 7 6
## robinson 7 7 7 9 6 8 10 6 7
## sebestyen 8 10 12 8 7 6 12 8 8
## kettunen 9 9 13 6 13 10 7 11 14
## volchkova 10 6 14 11 10 12 6 9 15
## maniachenko 13 12 11 12 16 11 11 10 9
## fontana 14 11 18 16 9 15 9 12 10
## liashenko 15 13 6 10 8 14 13 14 16
## onda 11 14 10 15 15 13 15 13 11
## hubert 12 17 17 13 11 16 14 15 13
## meier 16 16 9 14 14 9 16 16 12
## gusmeroli 17 15 15 17 17 18 17 17 17
## soldatova 19 18 22 20 21 17 18 18 19
## hegel 20 21 16 22 18 19 21 19 18
## giunchi 18 19 20 21 19 20 20 20 20
## babiakova 22 20 19 19 20 21 19 22 22
## kopac 21 22 23 18 22 22 22 21 21
## luca 23 23 21 23 23 23 23 23 23We might ask how judges J1 and J2 compare
to one another? We need to create vectors like x and
y above:
j1 <- unclass(skating_table)[,1] # column 1 is judge number 1
names(j1) <- rownames(skating_table)
j2 <- unclass(skating_table)[,2] # column 2 is judge number 2
names(j2) <- rownames(skating_table)
j1## hughes slutskaya kwan cohen suguri butyrskaya
## 1 3 2 5 4 6
## robinson sebestyen kettunen volchkova maniachenko fontana
## 7 8 9 10 13 14
## liashenko onda hubert meier gusmeroli soldatova
## 15 11 12 16 17 19
## hegel giunchi babiakova kopac luca
## 20 18 22 21 23
j2## hughes slutskaya kwan cohen suguri butyrskaya
## 4 1 3 2 8 5
## robinson sebestyen kettunen volchkova maniachenko fontana
## 7 10 9 6 12 11
## liashenko onda hubert meier gusmeroli soldatova
## 13 14 17 16 15 18
## hegel giunchi babiakova kopac luca
## 21 19 20 22 23
cbind(j1,j2)## j1 j2
## hughes 1 4
## slutskaya 3 1
## kwan 2 3
## cohen 5 2
## suguri 4 8
## butyrskaya 6 5
## robinson 7 7
## sebestyen 8 10
## kettunen 9 9
## volchkova 10 6
## maniachenko 13 12
## fontana 14 11
## liashenko 15 13
## onda 11 14
## hubert 12 17
## meier 16 16
## gusmeroli 17 15
## soldatova 19 18
## hegel 20 21
## giunchi 18 19
## babiakova 22 20
## kopac 21 22
## luca 23 23In the above, see how objects j1 and j2
have identical names, in the same order. Observe that
hughes is ranked 1 (that is, first) by J1, and
4 (that is, 4th) by J2. This makes it sensible to plot
j1 against j2:
par(pty='s') # forces plot to be square
plot(j1,j2,asp=1,pty='s',xlim=c(0,25),ylim=c(0,25),pch=16,xlab='judge 1',ylab='judge 2')
abline(0,1) # diagonal line
for(i in seq_along(j1)){text(j1[i],j2[i],names(j1)[i],pos=4,col='gray',cex=0.7)}
Judge 1 vs judge 2
In figure @ref(fig:j1vsj2), we see general agreement but differences
in detail. For example, hughes is ranked first by judge 1
and fourth by judge 2. However, other problems are not so easy. Suppose
we wish to compare the ranks according to likelihood with the ranks
according to some points system.
mL <- skating_maxp # predefined; use maxp(skating) to calculate ab initio
mL## babiakova butyrskaya cohen fontana giunchi gusmeroli
## 5.047235e-06 5.304470e-03 9.024149e-02 3.683282e-04 1.137326e-05 8.316914e-05
## hegel hubert hughes kettunen kopac kwan
## 9.785631e-06 2.689831e-04 2.983166e-01 1.376412e-03 2.996667e-06 2.655059e-01
## liashenko luca maniachenko meier onda robinson
## 5.188626e-04 1.000000e-06 8.068792e-04 2.928089e-04 4.880190e-04 5.308262e-03
## sebestyen slutskaya soldatova suguri volchkova
## 2.588302e-03 3.088930e-01 1.269275e-05 1.845715e-02 1.138379e-03Note that in the above, the competitors’ names are in alphabetical order. We first need to convert strengths to ranks:
mL[] <- rank(-mL) # minus because ranks orders from weak to strong
mL## babiakova butyrskaya cohen fontana giunchi gusmeroli
## 21 7 4 14 19 17
## hegel hubert hughes kettunen kopac kwan
## 20 16 2 9 22 3
## liashenko luca maniachenko meier onda robinson
## 12 23 11 15 13 6
## sebestyen slutskaya soldatova suguri volchkova
## 8 1 18 5 10(note that the names are in the same order as before, alphabetical).
In the above we see that slutskya ranks first,
hughes second, and so on. Another way of ranking the
skaters is to use a Borda-type system: essentially the rowsums of the
table (and, of course, the lowest score wins):
## hughes slutskaya kwan cohen suguri butyrskaya
## 1 2 3 4 5 6
## robinson sebestyen kettunen volchkova maniachenko fontana
## 7 8 9 10 11 13
## liashenko onda hubert meier gusmeroli soldatova
## 12 14 16 15 17 18
## hegel giunchi babiakova kopac luca
## 19 20 21 22 23It is not at all obvious how to compare mP and
mL. For example, we might be interested in
hegel. It takes some effort to find that her likelihood
rank is 20 and her Borda rank is 19. Function ordertrans()
facilitates this:
ordertrans(mP,names(mL)) ## babiakova butyrskaya cohen fontana giunchi gusmeroli
## 21 6 4 13 20 17
## hegel hubert hughes kettunen kopac kwan
## 19 16 1 9 22 3
## liashenko luca maniachenko meier onda robinson
## 12 23 11 15 14 7
## sebestyen slutskaya soldatova suguri volchkova
## 8 2 18 5 10See above how ordertrans() shows the points-based ranks
but in alphabetical order, to facilitate comparison with
mL. We can now plot these against one another:
plot(mL,ordertrans(mP,names(mL)))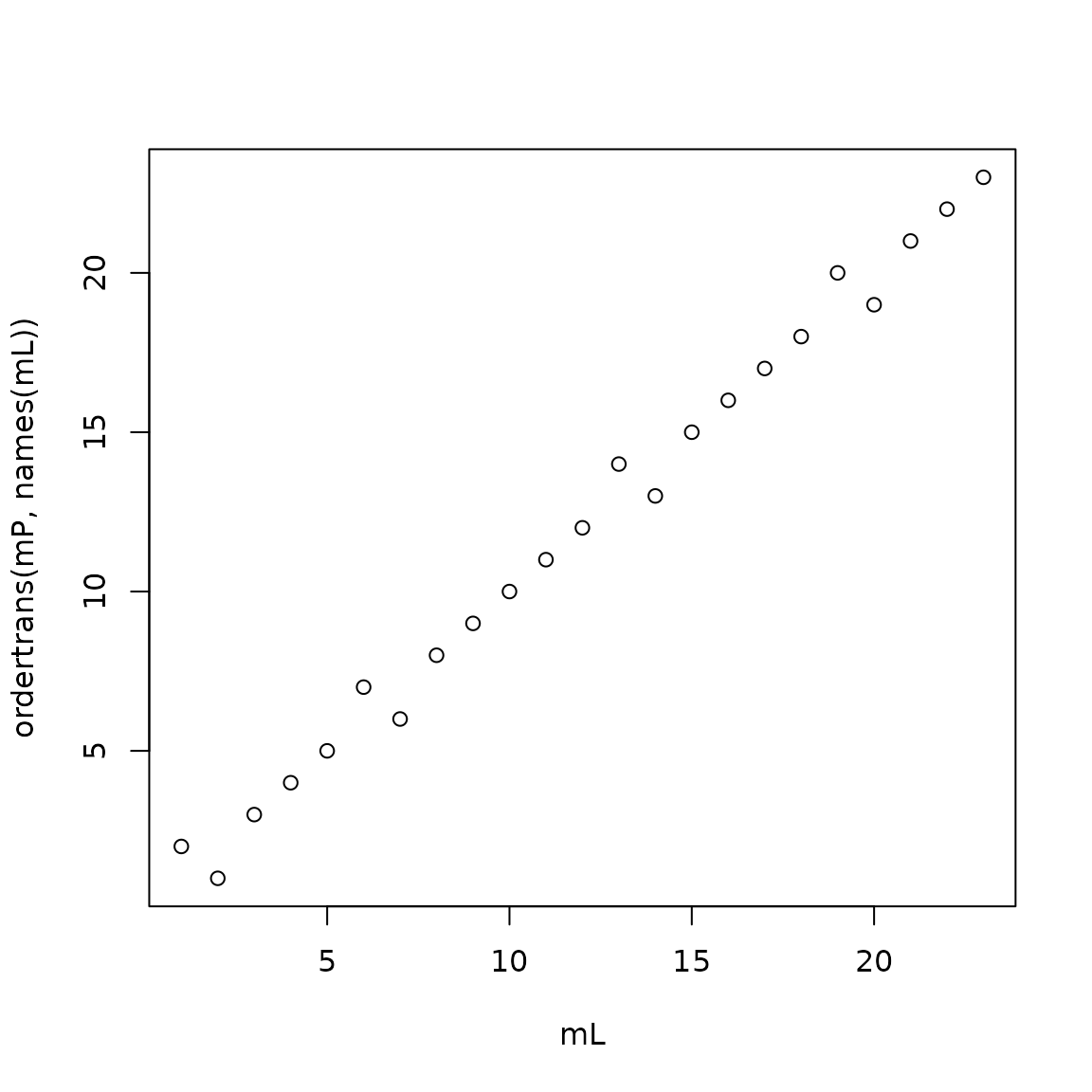
points-based ranks vs likelihood ranks
However, figure @ref(fig:crapplot) is a bit crude. Function
ordertransplot() gives a more visually pleasing output, see
figure @ref(fig:showoffordertransplot).
ordertransplot(mL,mP,xlab="likelihood rank",ylab="Borda rank")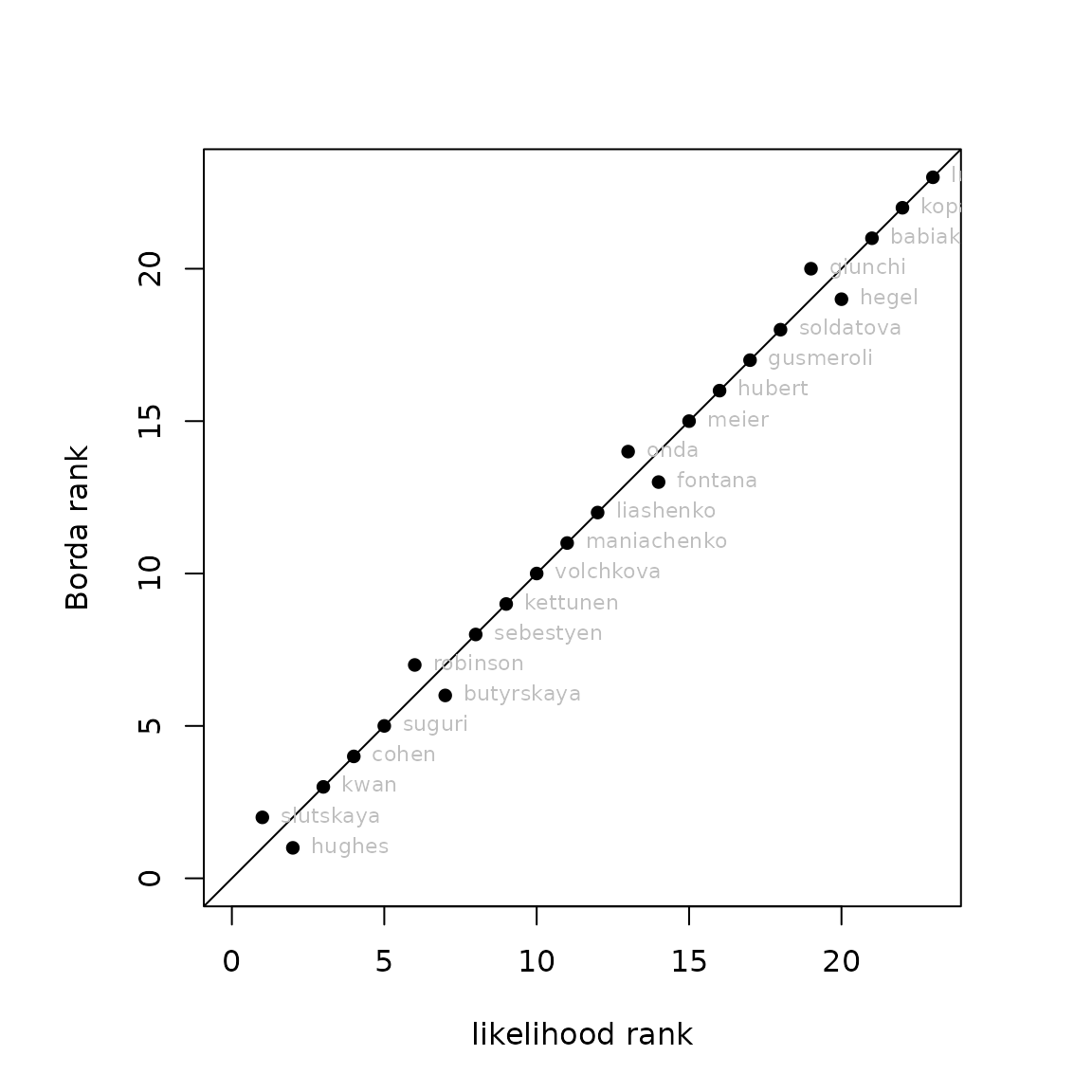
points=based rank vs likelihood rank using ordertransplot()
So now we may compare judge 1 against likelihood:
ordertransplot(mL,j1,xlab="likelihood rank",ylab="Judge 1 rank")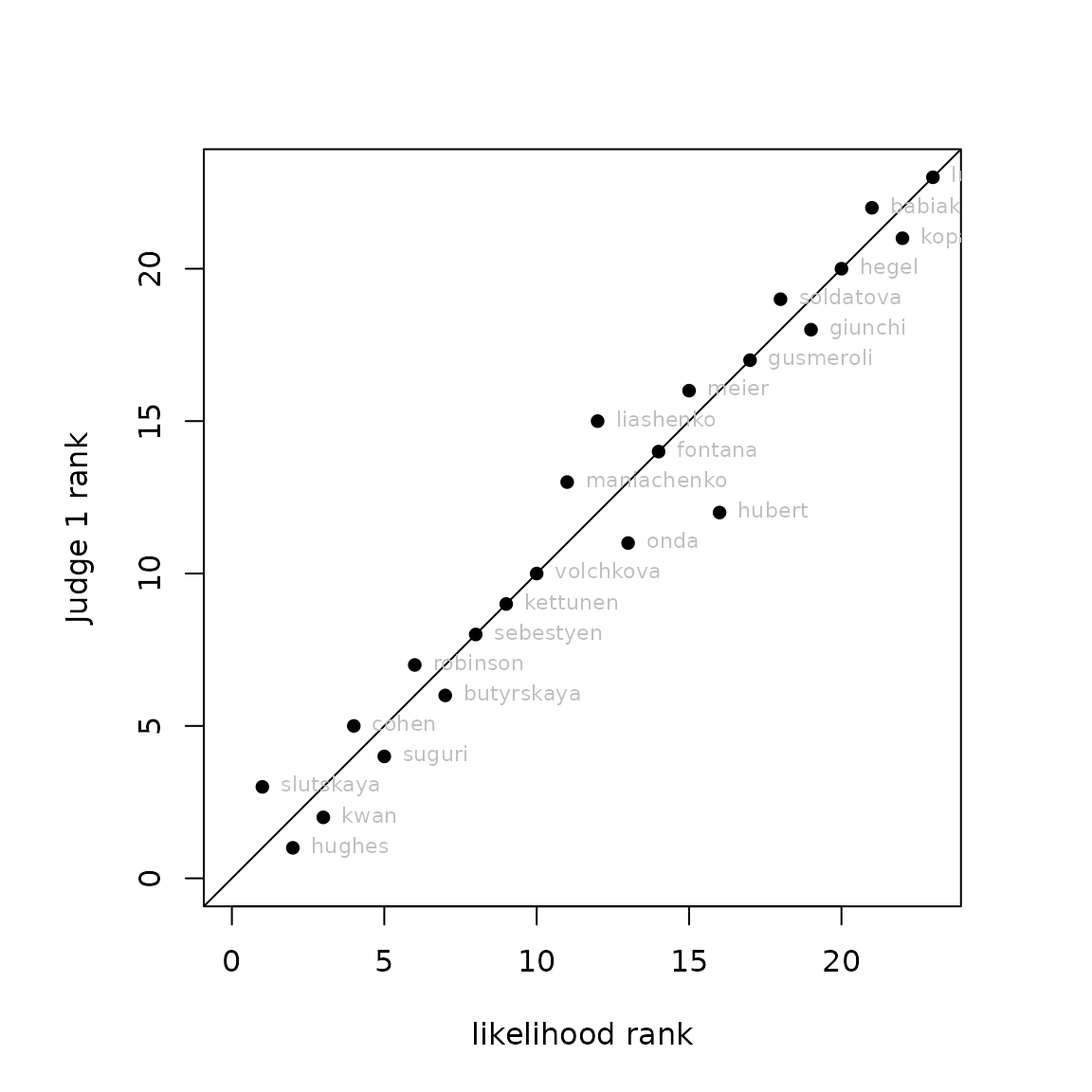
Likelihood rank vs rank according to Judge 1
In figure @ref(fig:transplotjudge1), looking at the lower-left corner, we see (reading horizontally) that the likelihood method placed Slutskya first, then Hughes second, then Kwan third; while (reading vertically) judge 1 placed Hughes first, then Kwan, then Slutskya.